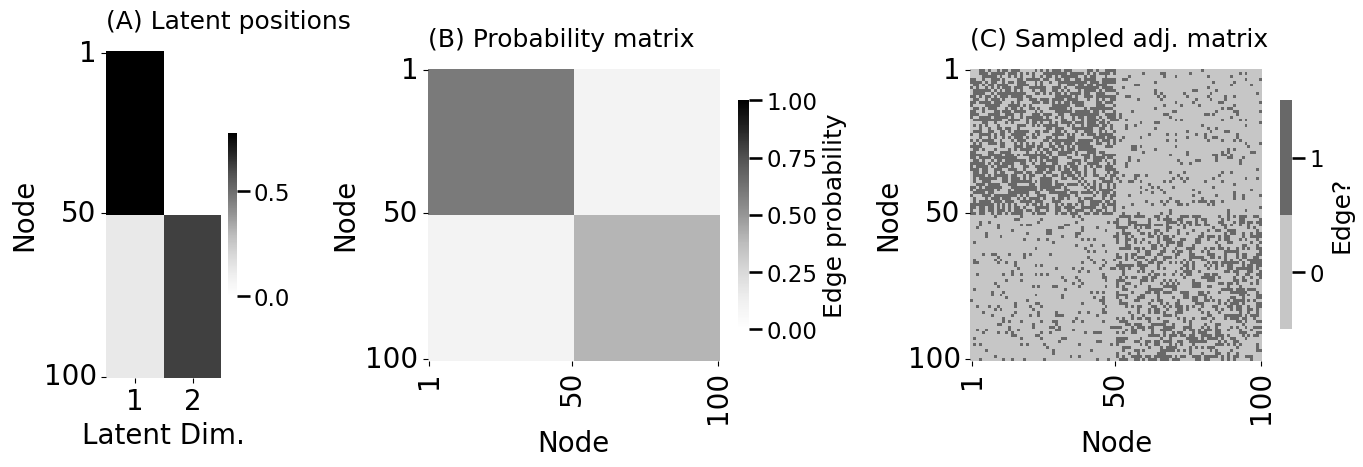
5.3 Adjacency spectral embedding#
mode = "svg"
import matplotlib
font = {'family' : 'Dejavu Sans',
'weight' : 'normal',
'size' : 20}
matplotlib.rc('font', **font)
import matplotlib
from matplotlib import pyplot as plt
from graspologic.simulations import sbm
from graphbook_code import generate_sbm_pmtx, lpm_from_sbm
import numpy as np
n = 100
# construct the block matrix B as described above
B = np.array([[0.6, 0.1],
[0.1, 0.4]])
# sample a graph from SBM_{100}(tau, B)
np.random.seed(0)
A, zs = sbm(n=[n//2, n//2], p=B, return_labels=True)
zs = zs + 1 # shift to 1-index
X = lpm_from_sbm(zs, B)
P = generate_sbm_pmtx(zs, B)
from graphbook_code import lpm_heatmap, heatmap
import os
fig, axs = plt.subplots(1, 3, figsize=(14, 5), gridspec_kw={"width_ratios": [.5, 1.27, 1.27]})
lpm_heatmap(X, xtitle="Latent Dim.", xticks=[0.5, 1.5], xticklabels=[1, 2],
yticks=[0.5, 49.5, 99.5], yticklabels=[1, 50, 100], ytitle="Node",
ax=axs[0], title="(A) Latent positions")
heatmap(P, legend_title="Edge probability", ax=axs[1],
xticks=[0.5, 49.5, 99.5], xticklabels=[1, 50, 100], xtitle="Node",
yticks=[0.5, 49.5, 99.5], yticklabels=[1, 50, 100], ytitle="Node",
title="(B) Probability matrix", vmin=0, vmax=1)
heatmap(A.astype(int), legend_title="Edge?", ax=axs[2],
xticks=[0.5, 49.5, 99.5], xticklabels=[1, 50, 100], xtitle="Node",
yticks=[0.5, 49.5, 99.5], yticklabels=[1, 50, 100], ytitle="Node",
title="(C) Sampled adj. matrix")
fig.tight_layout()
os.makedirs("Figures", exist_ok=True)
fname = "ase_sbm_ex"
if mode != "png":
os.makedirs(f"Figures/{mode:s}", exist_ok=True)
fig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
os.makedirs("Figures/png", exist_ok=True)
fig.savefig(f"Figures/png/{fname:s}.png")
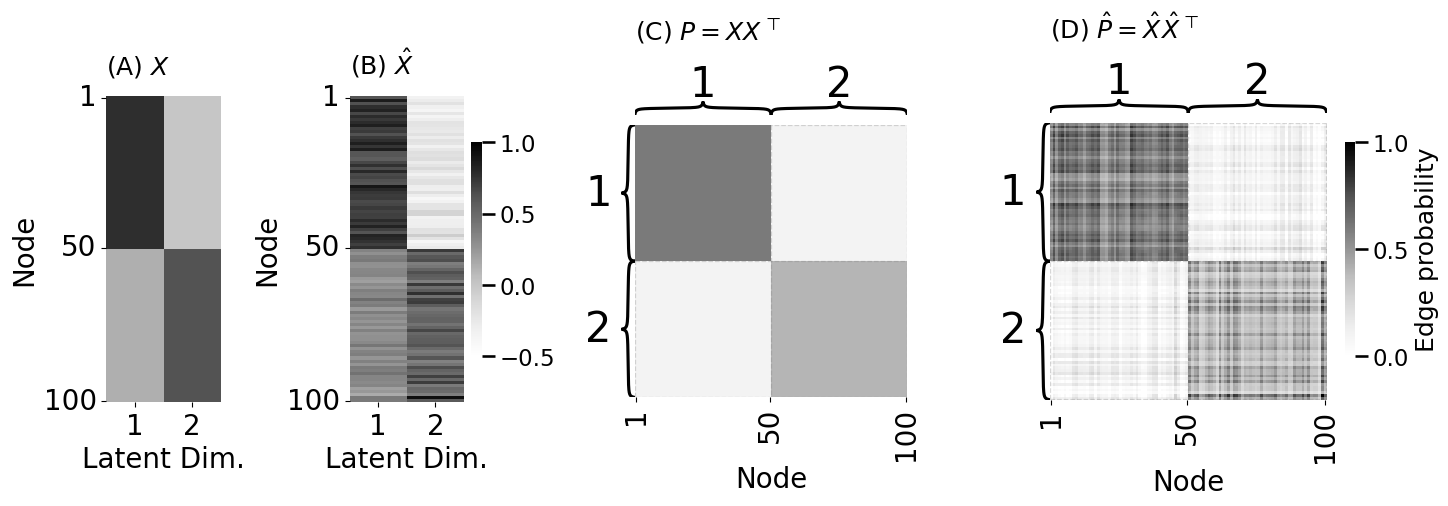
from graspologic.embed import AdjacencySpectralEmbed as ase
d = 2 # the latent dimensionality
# estimate the latent position matrix with ase
Xhat = ase(n_components=d, svd_seed=0).fit_transform(A)
Phat = Xhat @ Xhat.transpose()
fig, axs = plt.subplots(1, 4, figsize=(15, 5), gridspec_kw={"width_ratios": [.4, .5, 1, 1.27]})
lpm_heatmap(X, xtitle="Latent Dim.", xticks=[0.5, 1.5], xticklabels=[1, 2],
yticks=[0.5, 49.5, 99.5], cbar=False, yticklabels=[1, 50, 100], ytitle="Node",
ax=axs[0], title="(A) $X$", vmin=-.5, vmax=1)
lpm_heatmap(Xhat, xtitle="Latent Dim.", xticks=[0.5, 1.5], xticklabels=[1, 2],
yticks=[0.5, 49.5, 99.5], yticklabels=[1, 50, 100], ytitle="Node",
ax=axs[1], title="(B) $\hat X$", shrink=0.7, vmin=-.5, vmax=1)
heatmap(P, cbar=False, ax=axs[2],
xticks=[0.5, 49.5, 99.5], xticklabels=[1, 50, 100], xtitle="Node",
inner_hier_labels=zs, title="(C) $P = XX^\\top$", vmin=0, vmax=1)
heatmap(Phat, legend_title="Edge probability", ax=axs[3],
xticks=[0.5, 49.5, 99.5], xticklabels=[1, 50, 100], xtitle="Node",
inner_hier_labels=zs, title="(D) $\hat P = \\hat X \\hat X^\\top$", vmin=0, vmax=1)
fig.tight_layout()
fname = "ase_result"
if mode != "png":
fig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
fig.savefig(f"Figures/png/{fname:s}.png")
<>:9: SyntaxWarning: invalid escape sequence '\h'
<>:16: SyntaxWarning: invalid escape sequence '\h'
<>:9: SyntaxWarning: invalid escape sequence '\h'
<>:16: SyntaxWarning: invalid escape sequence '\h'
/tmp/ipykernel_4201/638704847.py:9: SyntaxWarning: invalid escape sequence '\h'
ax=axs[1], title="(B) $\hat X$", shrink=0.7, vmin=-.5, vmax=1)
/tmp/ipykernel_4201/638704847.py:16: SyntaxWarning: invalid escape sequence '\h'
inner_hier_labels=zs, title="(D) $\hat P = \\hat X \\hat X^\\top$", vmin=0, vmax=1)
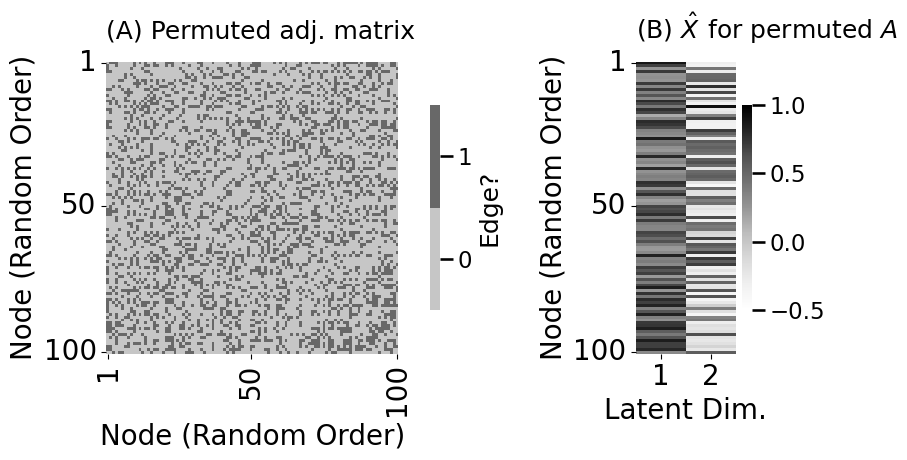
vtx_perm = np.random.choice(n, size=n, replace=False)
# reorder the adjacency matrix
Aperm = A[tuple([vtx_perm])] [:,vtx_perm]
# reorder the community assignment vector
zperm = np.array(zs)[vtx_perm]
# compute the estimated latent positions using the
# permuted adjacency matrix
Xhat_perm = ase(n_components=2, svd_seed=0).fit_transform(Aperm)
import matplotlib.gridspec as gridspec
import matplotlib.image as mpimg
fig, axs = plt.subplots(1, 2, figsize=(10, 5), gridspec_kw={"width_ratios": [1, .2]})
heatmap(Aperm.astype(int), legend_title="Edge?", ax=axs[0],
xticks=[0.5, 49.5, 99.5], xticklabels=[1, 50, 100], xtitle="Node (Random Order)",
yticks=[0.5, 49.5, 99.5], yticklabels=[1, 50, 100], ytitle="Node (Random Order)",
title="(A) Permuted adj. matrix")
lpm_heatmap(Xhat_perm, xtitle="Latent Dim.", xticks=[0.5, 1.5], xticklabels=[1, 2],
yticks=[0.5, 49.5, 99.5], yticklabels=[1, 50, 100], ytitle="Node (Random Order)",
ax=axs[1], title="(B) $\hat X$ for permuted $A$", shrink=0.7, vmin=-.5, vmax=1)
fig.tight_layout()
fname = "ase_permutedab"
if mode != "png":
fig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
fig.savefig(f"Figures/png/{fname:s}.png")
<>:12: SyntaxWarning: invalid escape sequence '\h'
<>:12: SyntaxWarning: invalid escape sequence '\h'
/tmp/ipykernel_4201/2433709469.py:12: SyntaxWarning: invalid escape sequence '\h'
ax=axs[1], title="(B) $\hat X$ for permuted $A$", shrink=0.7, vmin=-.5, vmax=1)
import pandas as pd
def tidy_lpm(X, labels=None, label_name="Community"):
"""
A function which takes a latent position matrix X,
and tidyfies it for seaborn. Optionally, adds a column
for the community of the nodes.
"""
X_tidy = pd.DataFrame(X)
X_tidy.columns = ["Dimension {:d}".format(d + 1) for d in range(0, X.shape[1])]
if labels is not None:
X_tidy[label_name] = labels
return X_tidy
Xhat_perm_tidy = tidy_lpm(Xhat_perm, labels=zperm)
import seaborn as sns
def pairplot(X, labels=None, label_name="Community", title=None, **kwargs):
"""
A function which takes a latent position matrix X,
and plots it as a pairplot with seaborn.
"""
X_tidy = tidy_lpm(X, labels=labels, label_name=label_name)
cols_to_pairplot = [col for col in X_tidy.columns.tolist() if "Dimension" in col]
fig = sns.pairplot(Xhat_perm_tidy, vars=["Dimension 1", "Dimension 2"], **kwargs)
# delete useless axis values
for ax in fig.axes.flatten():
ax.set_yticks([])
ax.set_xticks([])
if title is not None:
fig.fig.suptitle(title, y=1.05)
if labels is not None and "hue" in kwargs.keys():
for lh in fig._legend.legend_handles:
lh.set_alpha(1)
lh._sizes = [100]
return fig
fig = pairplot(Xhat_perm, labels=zperm, diag_kind="hist", title="(C) Pairs plot of $\\hat X$ for permuted $A$", height=2.5,
plot_kws={"color": "#000000"}, diag_kws={"color": "#000000"})
fig.tight_layout()
fname = "ase_permutedc"
if mode != "png":
fig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
fig.savefig(f"Figures/png/{fname:s}.png")
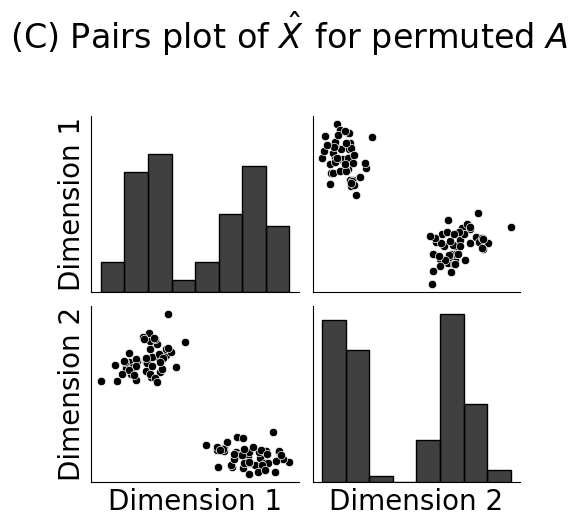
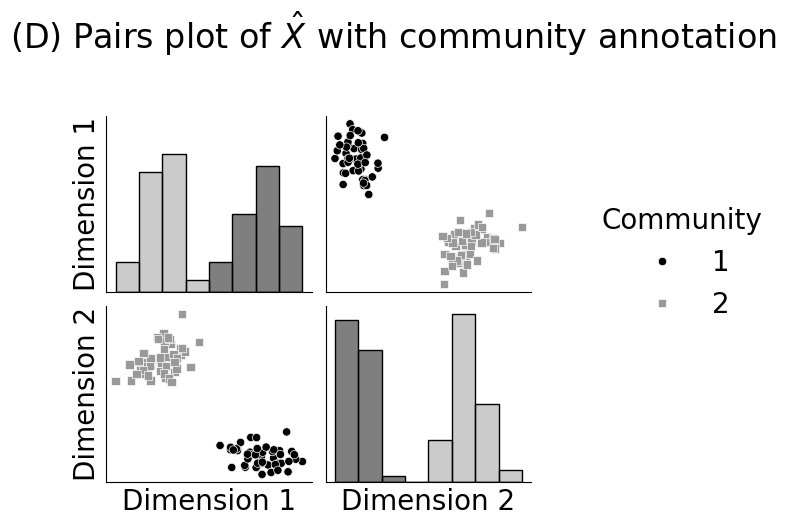
fig = pairplot(Xhat_perm, labels=zperm, diag_kind="hist", title="(D) Pairs plot of $\\hat X$ with community annotation", height=2.5,
hue="Community", palette={1: "#000000", 2: "#999999"}, markers={1: "o", 2: "s"})
fig.tight_layout()
fname = "ase_permutedd"
if mode != "png":
fig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
fig.savefig(f"Figures/png/{fname:s}.png")
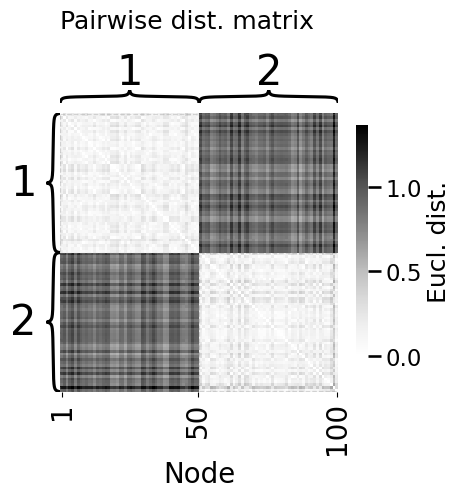
from scipy.spatial import distance_matrix
D = distance_matrix(Xhat, Xhat)
import math
from matplotlib import patches
fig, axs = plt.subplots(1, 1, figsize=(5, 5))
heatmap(D, title="Pairwise dist. matrix", ax=axs,
xticks=[0.5, 49.5, 99.5], xticklabels=[1, 50, 100], xtitle="Node",
inner_hier_labels=zs, legend_title="Eucl. dist.")
fig.tight_layout()
fname = "dist"
if mode != "png":
fig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
fig.savefig(f"Figures/png/{fname:s}.png")