import os
import numpy as np
from graphbook_code import heatmap
from graspologic.simulations import sample_edges
def generate_unit_circle(radius):
diameter = 2*radius + 1
rx = ry = diameter/2
x, y = np.indices((diameter, diameter))
circle_dist = np.hypot(rx - x, ry - y)
diff_from_radius = np.abs(circle_dist - radius)
less_than_half = diff_from_radius < 0.5
return less_than_half.astype(int)
def add_smile():
canvas = np.zeros((51, 51))
canvas[2:45, 2:45] = generate_unit_circle(21)
mask = np.zeros((51, 51), dtype=bool)
mask[np.triu_indices_from(mask)] = True
upper_left = np.rot90(mask)
canvas[upper_left] = 0
return canvas
def smile_probability(upper_p, lower_p):
smiley = add_smile()
P = generate_unit_circle(25)
P[5:16, 25:36] = generate_unit_circle(5)
P[smiley != 0] = smiley[smiley != 0]
mask = np.zeros((51, 51), dtype=bool)
mask[np.triu_indices_from(mask)] = True
P[~mask] = 0
# symmetrize the probability matrix
P = (P + P.T - np.diag(np.diag(P))).astype(float)
P[P == 1] = lower_p
P[P == 0] = upper_p
return P
fig, axs = plt.subplots(1, 2, figsize=(12, 5))
P = smile_probability(.95, 0.05)
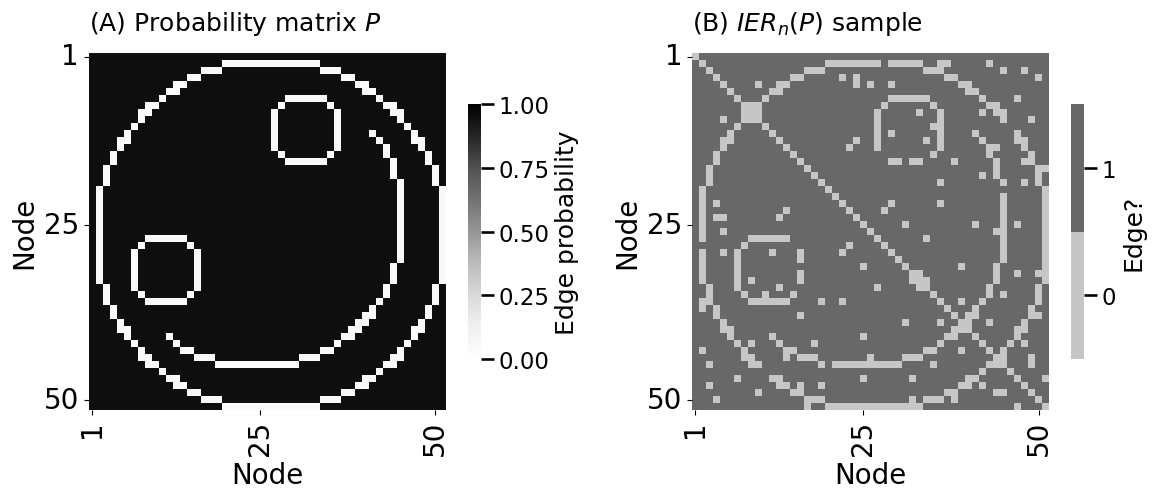
heatmap(P, vmin=0, vmax=1, title="(A) Probability matrix $P$",
legend_title="Edge probability", xtitle="Node", ytitle="Node",
xticks=[0.5, 24.5, 49.5], xticklabels=[1,25,50],
yticks=[0.5, 24.5, 49.5], yticklabels=[1,25,50],
ax=axs[0])
A = sample_edges(P, directed=False, loops=False)
heatmap(A.astype(int), vmin=0, vmax=1, title="(B) $IER_n(P)$ sample",
legend_title="Edge?", xtitle="Node", ytitle="Node",
xticks=[0.5, 24.5, 49.5], xticklabels=[1,25,50],
yticks=[0.5, 24.5, 49.5], yticklabels=[1,25,50],
ax=axs[1])
fig.tight_layout()
os.makedirs("Figures", exist_ok=True)
fname = "ier"
if mode != "png":
os.makedirs(f"Figures/{mode:s}", exist_ok=True)
fig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
os.makedirs("Figures/png", exist_ok=True)
fig.savefig(f"Figures/png/{fname:s}.png")