4.5 Positive Semi-Definite Matrices#
mode = "svg"
import matplotlib
font = {'family' : 'Dejavu Sans',
'weight' : 'normal',
'size' : 20}
matplotlib.rc('font', **font)
import matplotlib
from matplotlib import pyplot as plt
import numpy as np
def block_mtx_psd(B):
"""
A function which indicates whether a matrix
B is positive semi-definite.
"""
return np.all(np.linalg.eigvals(B) >= 0)
import numpy as np
from graphbook_code import heatmap
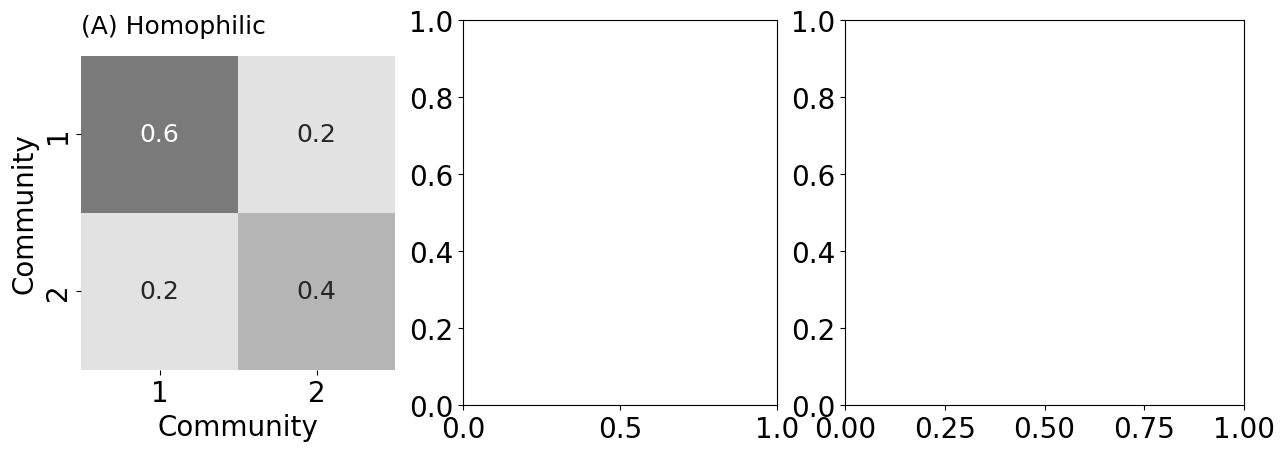
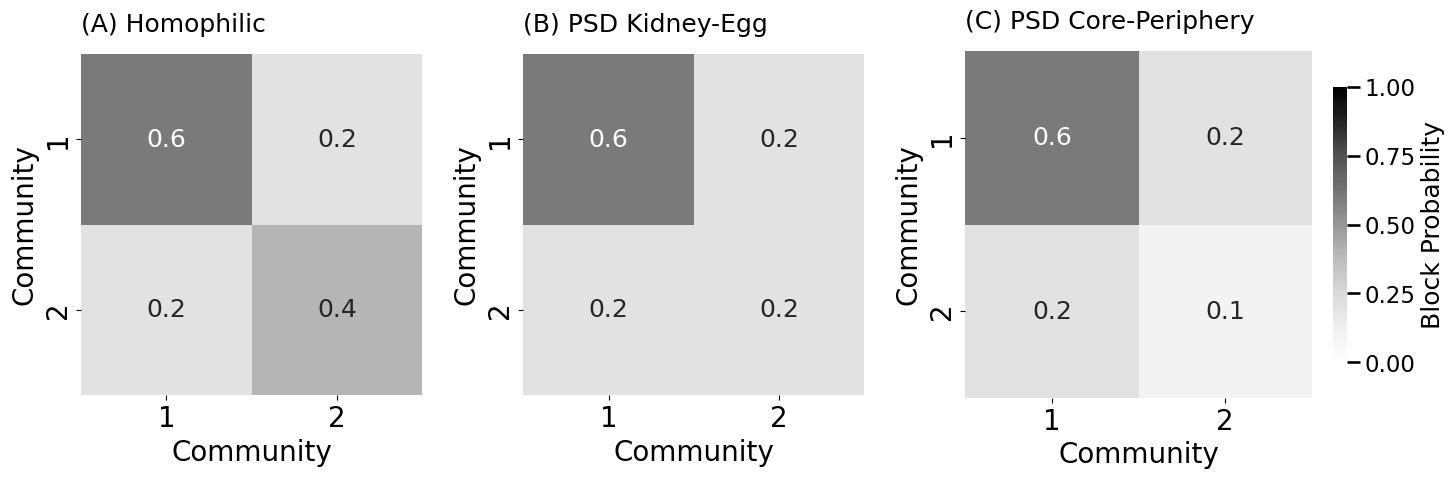
B = np.array([[0.6, 0.2],
[0.2, 0.4]])
block_mtx_psd(B)
# True
True
psdfig, psdaxs = plt.subplots(1, 3, figsize=(15, 5), gridspec_kw={"width_ratios": [1,1,1.27]})
heatmap(B, title="(A) Homophilic", ax=psdaxs[0],
xtitle="Community", ytitle="Community",
xticks=[0.5, 1.5], yticks=[0.5, 1.5],
xticklabels=[1, 2], yticklabels=[1, 2], vmin=0, vmax=1,
cbar=False, annot=True)
<Axes: title={'left': '(A) Homophilic'}, xlabel='Community', ylabel='Community'>
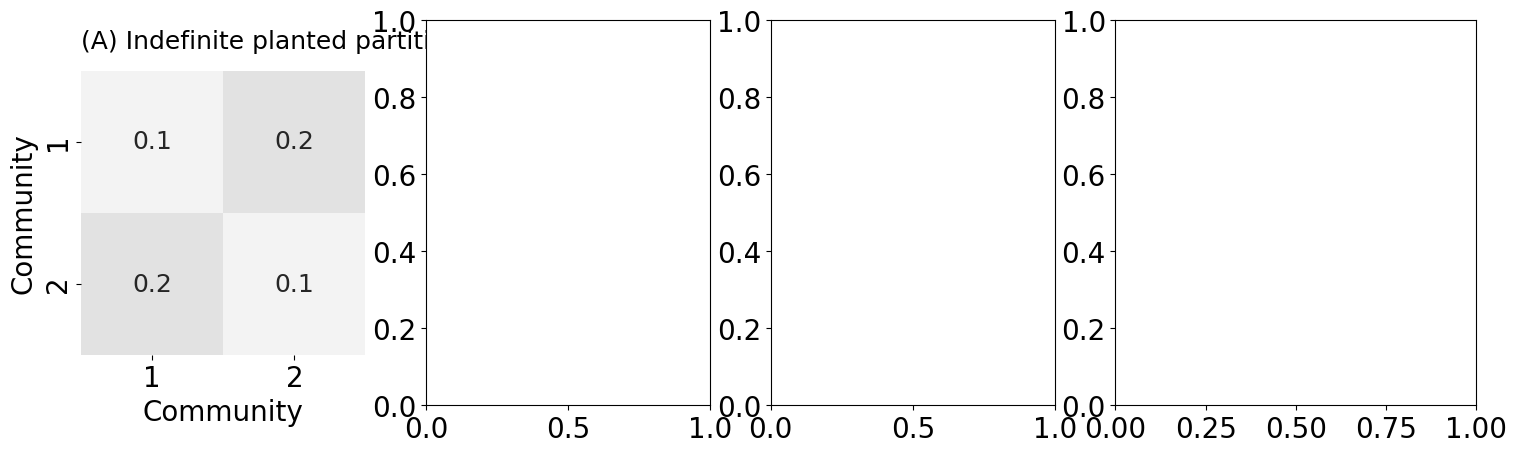
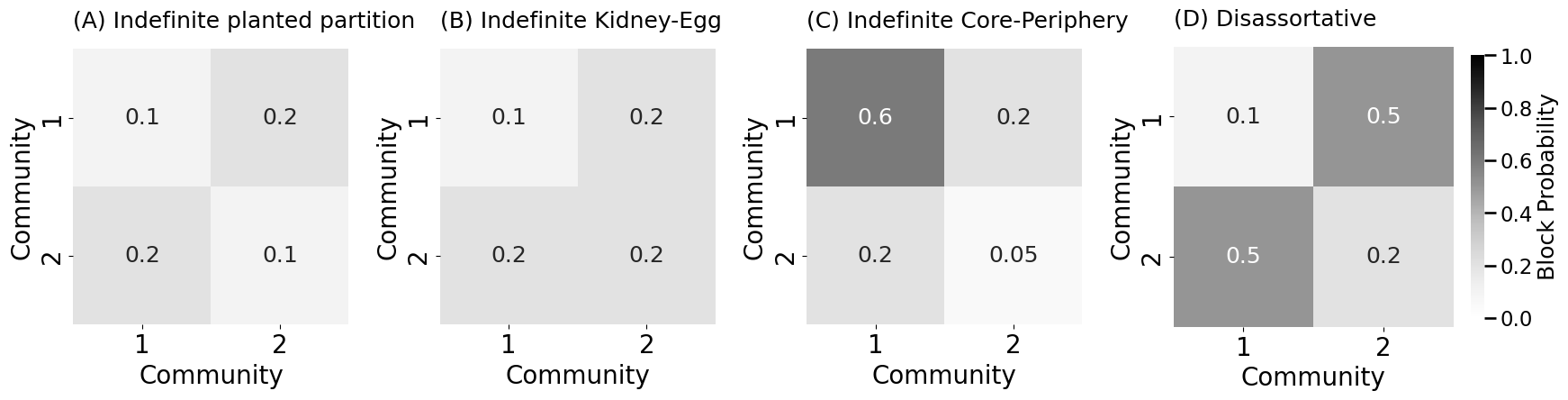
B_indef = np.array([[.1, .2],
[.2, .1]])
block_mtx_psd(B_indef)
# False
False
indeffig, indefaxs = plt.subplots(1, 4, figsize=(18, 5), gridspec_kw={"width_ratios": [1,1,1,1.27]})
heatmap(B_indef, title="(A) Indefinite planted partition", ax=indefaxs[0],
xtitle="Community", ytitle="Community",
xticks=[0.5, 1.5], yticks=[0.5, 1.5],
xticklabels=[1, 2], yticklabels=[1, 2], vmin=0, vmax=1,
cbar=False, annot=True)
<Axes: title={'left': '(A) Indefinite planted partition'}, xlabel='Community', ylabel='Community'>
# a positive semi-definite kidney-egg block matrix
B_psd = np.array([[.6, .2],
[.2, .2]])
block_mtx_psd(B_psd)
# True
True
heatmap(B_psd, title="(B) PSD Kidney-Egg", ax=psdaxs[1],
xtitle="Community", ytitle="Community",
xticks=[0.5, 1.5], yticks=[0.5, 1.5],
xticklabels=[1, 2], yticklabels=[1, 2], vmin=0, vmax=1,
cbar=False, annot=True)
<Axes: title={'left': '(B) PSD Kidney-Egg'}, xlabel='Community', ylabel='Community'>
# an indefinite kidney-egg block matrix
B_indef = np.array([[.1, .2],
[.2, .2]])
block_mtx_psd(B_indef)
#False
False
heatmap(B_indef, title="(B) Indefinite Kidney-Egg", ax=indefaxs[1],
xtitle="Community", ytitle="Community",
xticks=[0.5, 1.5], yticks=[0.5, 1.5],
xticklabels=[1, 2], yticklabels=[1, 2], vmin=0, vmax=1,
cbar=False, annot=True)
<Axes: title={'left': '(B) Indefinite Kidney-Egg'}, xlabel='Community', ylabel='Community'>
# a positive semi-definite core-periphery block matrix
B_psd = np.array([[.6, .2],
[.2, .1]])
block_mtx_psd(B_psd)
# True
True
heatmap(B_psd, title="(C) PSD Core-Periphery", ax=psdaxs[2],
xtitle="Community", ytitle="Community",
xticks=[0.5, 1.5], yticks=[0.5, 1.5],
xticklabels=[1, 2], yticklabels=[1, 2], vmin=0, vmax=1,
cbar=True, annot=True, legend_title="Block Probability")
<Axes: title={'left': '(C) PSD Core-Periphery'}, xlabel='Community', ylabel='Community'>
# an indefinite core-periphery block matrix
B_indef = np.array([[.6, .2],
[.2, .05]])
block_mtx_psd(B_indef)
# False
False
heatmap(B_indef, title="(C) Indefinite Core-Periphery", ax=indefaxs[2],
xtitle="Community", ytitle="Community",
xticks=[0.5, 1.5], yticks=[0.5, 1.5],
xticklabels=[1, 2], yticklabels=[1, 2], vmin=0, vmax=1,
cbar=False, annot=True)
<Axes: title={'left': '(C) Indefinite Core-Periphery'}, xlabel='Community', ylabel='Community'>
# an indefinite disassortative block matrix
B = np.array([[.1, .5],
[.5, .2]])
block_mtx_psd(B)
# False
False
heatmap(B, title="(D) Disassortative", ax=indefaxs[3],
xtitle="Community", ytitle="Community",
xticks=[0.5, 1.5], yticks=[0.5, 1.5],
xticklabels=[1, 2], yticklabels=[1, 2], vmin=0, vmax=1,
cbar=True, annot=True, legend_title="Block Probability")
<Axes: title={'left': '(D) Disassortative'}, xlabel='Community', ylabel='Community'>
import os
psdfig.tight_layout()
os.makedirs("Figures", exist_ok=True)
fname = "psd"
if mode != "png":
os.makedirs(f"Figures/{mode:s}", exist_ok=True)
psdfig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
os.makedirs("Figures/png", exist_ok=True)
psdfig.savefig(f"Figures/png/{fname:s}.png")
psdfig
indeffig.tight_layout()
fname = "indef"
if mode != "png":
indeffig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
indeffig.savefig(f"Figures/png/{fname:s}.png")
indeffig
# homophilic, and hence positive semi-definite, block matrix
B = np.array([[0.6, 0.2],
[0.2, 0.4]])
# generate square root matrix
sqrtB = np.linalg.cholesky(B)
# verify that the process worked through by equality element-wise
# use allclose instead of array_equal because of tiny
# numerical precision errors
np.allclose(sqrtB @ sqrtB.T, B)
# True
True
from graphbook_code import ohe_comm_vec
def lpm_from_sbm(z, B):
"""
A function to produce a latent position matrix from a
community assignment vector and a block matrix.
"""
if not block_mtx_psd(B):
raise ValueError("Latent position matrices require PSD block matrices!")
# one-hot encode the community assignment vector
C = ohe_comm_vec(z)
# compute square root matrix
sqrtB = np.linalg.cholesky(B)
# X = C*sqrt(B)
return C @ sqrtB
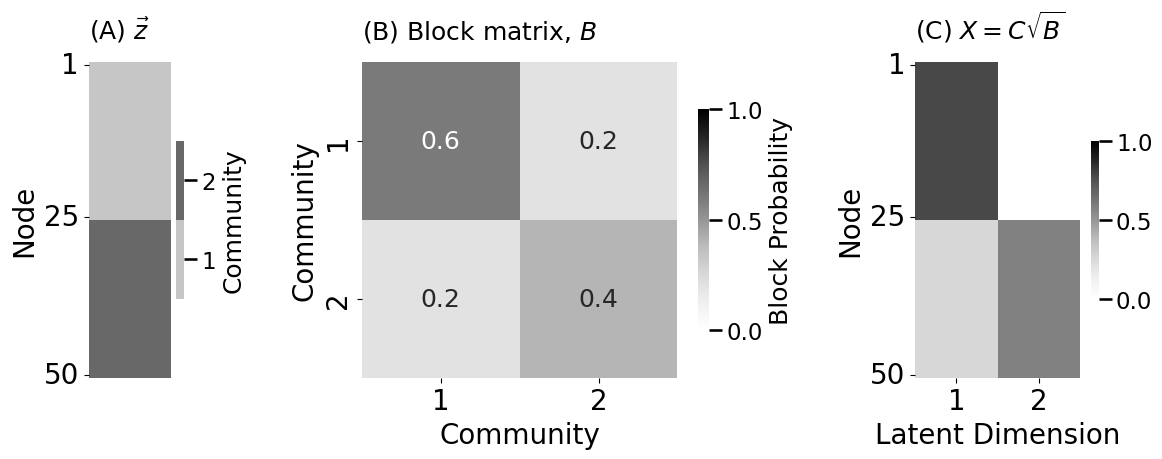
# make a community assignment vector for 25 nodes / community
nk = 25
z = np.repeat([1, 2], nk)
# latent position matrix for an equivalent RDPG
X = lpm_from_sbm(z, B)
from graphbook_code import plot_vector, lpm_heatmap
fig, axs = plt.subplots(1,3, figsize=(12, 5), gridspec_kw={"width_ratios": [.5,2,1]})
plot_vector(z.astype(int), title="(A) $\\vec z$", legend_title="Community",
ticks=[0.5, 24.5, 49.5], ticklabels=[1, 25, 50],
ticktitle="Node", ax=axs[0])
heatmap(B, title="(B) Block matrix, $B$", ax=axs[1],
xtitle="Community", ytitle="Community",
xticks=[0.5, 1.5], yticks=[0.5, 1.5],
xticklabels=[1, 2], yticklabels=[1, 2], vmin=0, vmax=1,
cbar=True, annot=True, legend_title="Block Probability")
lpm_heatmap(X, title="(C) $X = C\\sqrt{B}$", ax=axs[2],
xtitle="Latent Dimension", ytitle="Node",
yticks=[0.5, 24.5, 49.5], yticklabels=[1, 25, 50],
xticks=[0.5, 1.5], xticklabels=[1, 2], vmin=0, vmax=1,
cbar=True)
fig.tight_layout()
fname = "sbm_lpm"
if mode != "png":
fig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
fig.savefig(f"Figures/png/{fname:s}.png")
from graphbook_code import generate_sbm_pmtx
# generate the probability matrices for an RDPG using X and SBM
P_rdpg = X @ X.T
P_sbm = generate_sbm_pmtx(z, B)
# verify equality element-wise
np.allclose(P_rdpg, P_sbm)
# True
True
