9.1 Graph neural networks#
mode = "svg"
import matplotlib
font = {'family' : 'Dejavu Sans',
'weight' : 'normal',
'size' : 20}
matplotlib.rc('font', **font)
import matplotlib
from matplotlib import pyplot as plt
from torch_geometric.datasets import MoleculeNet
dataset = MoleculeNet(root='data/clintox', name='ClinTox')
print(f'Dataset: {dataset}\nNumber of molecules/graphs: {len(dataset)}\nNumber of classes: {dataset.num_classes}')
Downloading https://deepchemdata.s3-us-west-1.amazonaws.com/datasets/clintox.csv.gz
Extracting data/clintox/clintox/raw/clintox.csv.gz
Processing...
/opt/hostedtoolcache/Python/3.12.11/x64/lib/python3.12/site-packages/torch_geometric/datasets/molecule_net.py:213: UserWarning: Skipping molecule '[NH4][Pt]([NH4])(Cl)Cl' since it resulted in zero atoms
warnings.warn(f"Skipping molecule '{smiles}' since it "
/opt/hostedtoolcache/Python/3.12.11/x64/lib/python3.12/site-packages/torch_geometric/datasets/molecule_net.py:213: UserWarning: Skipping molecule 'c1ccc(cc1)n2c(=O)c(c(=O)n2c3ccccc3)CCS(=O)c4ccccc4' since it resulted in zero atoms
warnings.warn(f"Skipping molecule '{smiles}' since it "
/opt/hostedtoolcache/Python/3.12.11/x64/lib/python3.12/site-packages/torch_geometric/datasets/molecule_net.py:213: UserWarning: Skipping molecule 'CCCCc1c(=O)n(n(c1=O)c2ccc(cc2)O)c3ccccc3' since it resulted in zero atoms
warnings.warn(f"Skipping molecule '{smiles}' since it "
/opt/hostedtoolcache/Python/3.12.11/x64/lib/python3.12/site-packages/torch_geometric/datasets/molecule_net.py:213: UserWarning: Skipping molecule 'CCCCc1c(=O)n(n(c1=O)c2ccccc2)c3ccccc3' since it resulted in zero atoms
warnings.warn(f"Skipping molecule '{smiles}' since it "
Dataset: ClinTox(1480)
Number of molecules/graphs: 1480
Number of classes: 2
Done!
mols = dataset[26], dataset[83]
for m in mols:
print(m.smiles)
C([C@@H]1[C@H]([C@@H]([C@H](C(=O)O1)O)O)O)O
C1[C@@H]([C@H](O[C@H]1N2C=NC(=NC2=O)N)CO)O
from rdkit import Chem
from rdkit.Chem.Draw import rdMolDraw2D
from IPython.display import SVG
smiles = [Chem.MolFromSmiles(m.smiles) for m in mols]
d2d = rdMolDraw2D.MolDraw2DSVG(600,280,300,280)
d2d.drawOptions().addAtomIndices = True
d2d.DrawMolecules(smiles)
d2d.FinishDrawing()
SVG(d2d.GetDrawingText())
for i,m in enumerate(mols):
print(f'Molecule {i+1}: Number of atoms={m.x.shape[0]}, Features per atom={m.x.shape[1]}')
Molecule 1: Number of atoms=12, Features per atom=9
Molecule 2: Number of atoms=16, Features per atom=9
d2d = rdMolDraw2D.MolDraw2DSVG(600,280,300,280)
d2d.drawOptions().addBondIndices = True
d2d.DrawMolecules(smiles)
d2d.FinishDrawing()
SVG(d2d.GetDrawingText())
import numpy as np
_process = lambda x: [e[0] for e in np.split(x, 2)]
def adj_from_edgelist(molecule):
"""
A function that takes a molecule edgelist and produces an adjacency matrix.
"""
# the number of nodes is the number of atoms (rows of .x attribute)
n = molecule.x.shape[0]
# the adjacency matrix is n x n
A = np.zeros((n, n))
edgelist = m.edge_index.numpy()
# loop over the edges e_k, and for each edge, unpack the
# nodes that are incident it. for this pair of nodes,
# change the adjacency matrix entry to 1
for e_k, (i, j) in enumerate(zip(*_process(edgelist))):
A[i, j] = 1
return A
from graphbook_code import heatmap
for m_i, m in enumerate(mols):
A = adj_from_edgelist(m)
heatmap(A)
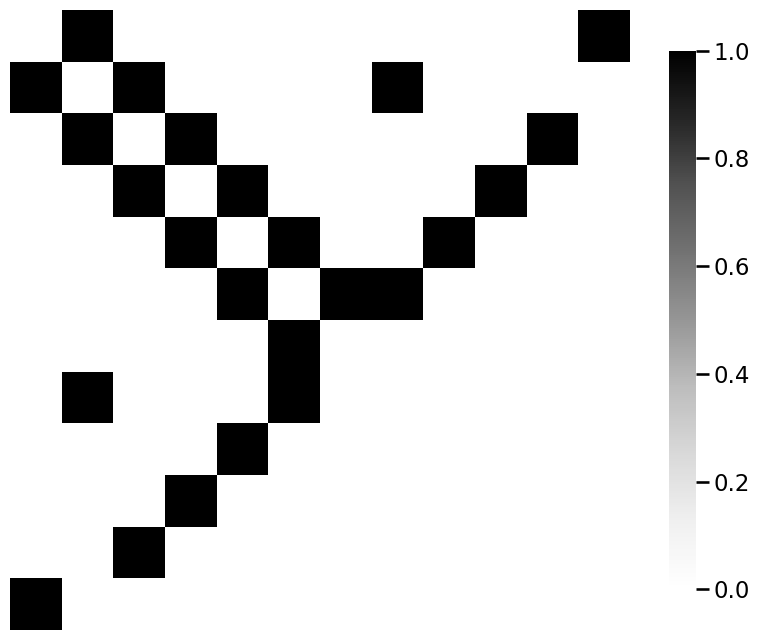
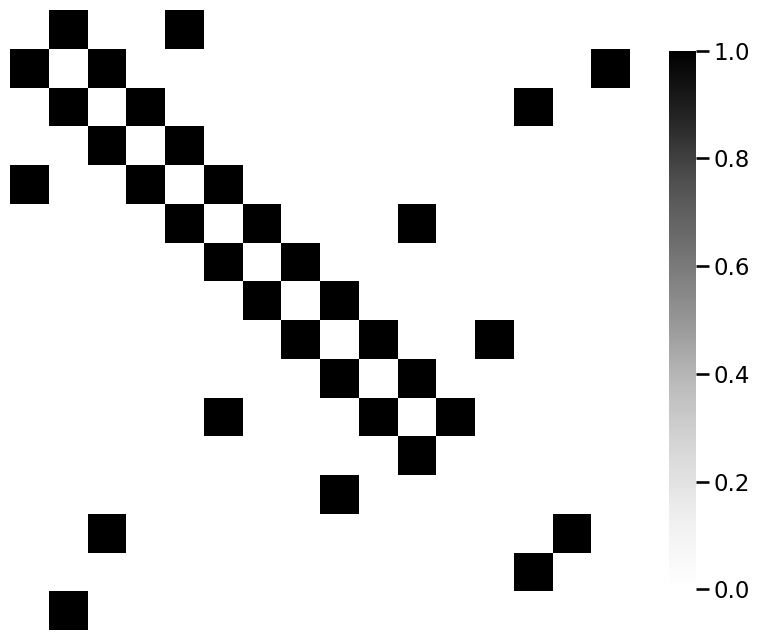
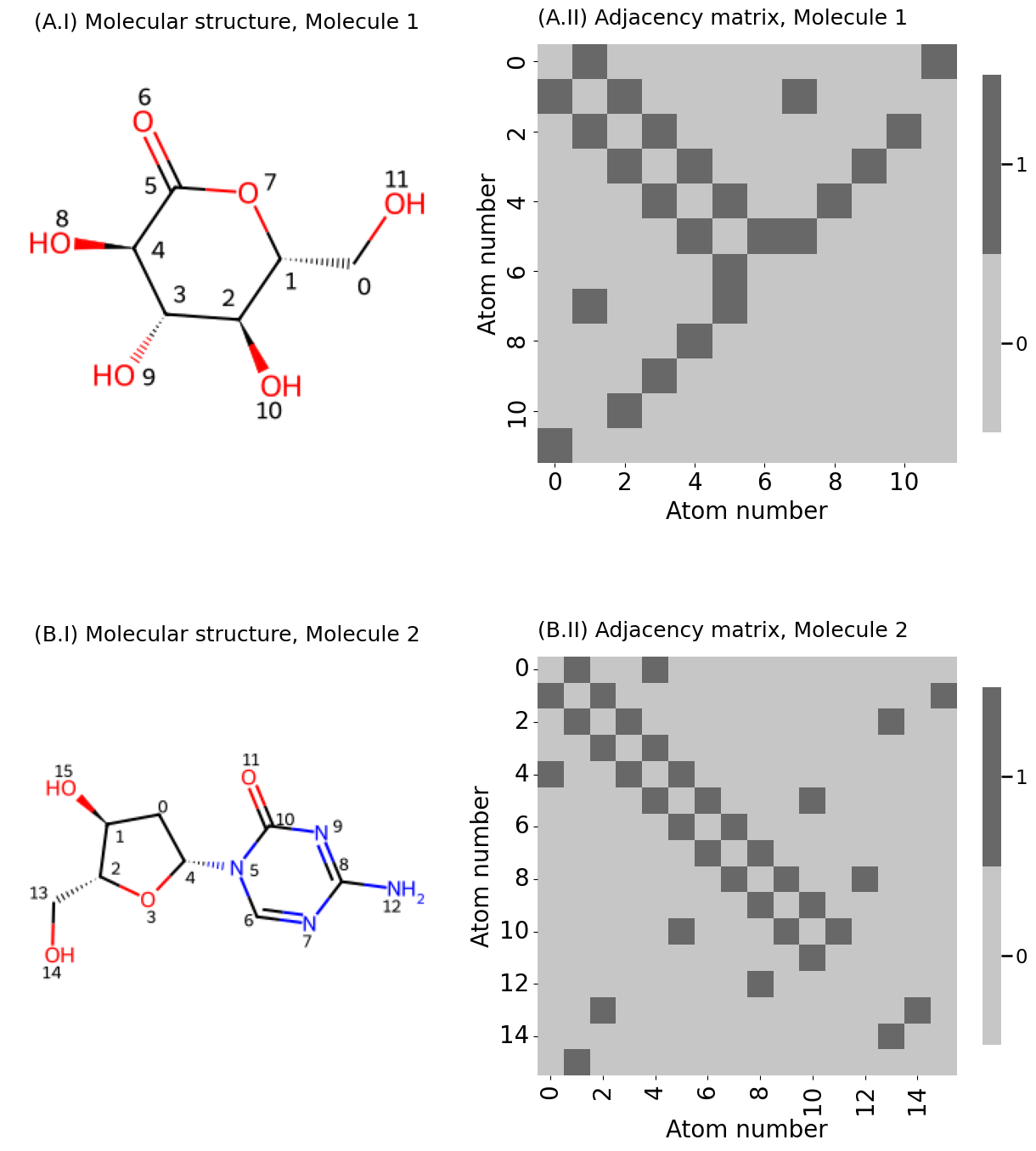
from PIL import Image
import os
fig, axs = plt.subplots(2, 2, figsize=(13, 15), gridspec_kw={"width_ratios": [1, 1.2]})
plot_titles = [[f"({x:s}.{y:s}" for y in [f"I) Molecular structure, {z:s}", f"II) Adjacency matrix, {z:s}"]] for x, z in zip(["A", "B"], ["Molecule 1", "Molecule 2"])]
title_left = ["Molecule 1", "Molecule 2"]
for i, m in enumerate(mols):
d2d = rdMolDraw2D.MolDraw2DCairo(300,300)
options = d2d.drawOptions()
options.addAtomIndices = True; options.minFontSize = 14; options.annotationFontScale = 0.8
d2d.DrawMolecule(Chem.MolFromSmiles(m.smiles))
d2d.FinishDrawing()
png_data = d2d.GetDrawingText()
# save png to file
png_fname = f'mol{i:d}.png'
with open(png_fname, 'wb') as png_file:
png_file.write(png_data)
axs[i][0].imshow(Image.open(png_fname))
axs[i][0].set_title(plot_titles[i][0], fontsize=18)
axs[i][0].axis("off")
A = adj_from_edgelist(m)
tick_range = range(0, np.ceil(A.shape[0] / 2).astype(int))
xticks = yticks = [2 * i + 0.5 for i in tick_range]
xticklabels = yticklabels = [f"{2 * i}" for i in tick_range]
heatmap(A.astype(int), ax=axs[i][1], xticks=xticks, xticklabels=xticklabels,
yticks=yticks, yticklabels=yticklabels, shrink=0.6,
title=plot_titles[i][1], xtitle="Atom number", ytitle="Atom number")
fig.tight_layout()
fname = "molecule_ex"
os.makedirs("Figures", exist_ok=True)
fname = "basic_mtxs"
if mode != "png":
os.makedirs(f"Figures/{mode:s}", exist_ok=True)
fig.savefig(f"Figures/{mode:s}/{fname:s}.{mode:s}")
os.makedirs("Figures/png", exist_ok=True)
fig.savefig(f"Figures/png/{fname:s}.png")
import torch
# for notebook reproducibility
torch.manual_seed(12345)
dataset = dataset.shuffle()
train_dataset = dataset[:1216]
test_dataset = dataset[1216:]
print(f'Number of training networks: {len(train_dataset)}')
print(f'Number of test networks: {len(test_dataset)}')
Number of training networks: 1216
Number of test networks: 264
from torch_geometric.loader import DataLoader
train_loader = DataLoader(train_dataset, batch_size=64, shuffle=True)
test_loader = DataLoader(test_dataset, batch_size=64, shuffle=False)
for step, data in enumerate(train_loader):
print(f'Step {step + 1}:')
print(f'Number of networks in the current batch: {data.num_graphs}')
print(data)
break
Step 1:
Number of networks in the current batch: 64
DataBatch(x=[1414, 9], edge_index=[2, 3014], edge_attr=[3014, 3], smiles=[64], y=[64, 2], batch=[1414], ptr=[65])
from torch.nn import Linear
import torch.nn.functional as F
from torch_geometric.nn import GCNConv
from torch_geometric.nn import global_mean_pool
torch.manual_seed(12345)
class GCN(torch.nn.Module):
def __init__(self, hidden_channels):
super(GCN, self).__init__()
self.conv1 = GCNConv(dataset.num_node_features, hidden_channels)
self.conv2 = GCNConv(hidden_channels, hidden_channels)
self.conv3 = GCNConv(hidden_channels, hidden_channels)
self.lin = Linear(hidden_channels, dataset.num_classes, bias=False)
def forward(self, x, edge_index, batch):
# 1. Obtain node embeddings via convolutional layers
x = self.conv1(x, edge_index)
x = x.relu()
x = self.conv2(x, edge_index)
x = x.relu()
x = self.conv3(x, edge_index)
# 2. Readout layer to produce network embedding
x = global_mean_pool(x, batch) # [batch_size, hidden_channels]
# 3. Apply a prediction classifier to the network embedding
x = self.lin(x)
return x
model = GCN(hidden_channels=64)
print(model)
GCN(
(conv1): GCNConv(9, 64)
(conv2): GCNConv(64, 64)
(conv3): GCNConv(64, 64)
(lin): Linear(in_features=64, out_features=2, bias=False)
)
model = GCN(hidden_channels=64)
optimizer = torch.optim.Adam(model.parameters(), lr=0.0001)
criterion = torch.nn.CrossEntropyLoss()
def train():
model.train()
for data in train_loader: # Iterate in batches over the training dataset.
out = model(data.x.float(), data.edge_index, data.batch) # Perform a single forward pass.
# Handle a pyg bug where last element in batch may be all zeros and excluded in the model output.
# https://github.com/pyg-team/pytorch_geometric/issues/1813
num_batch = out.shape[0]
loss = criterion(out, data.y[:num_batch, 0].long()) # Compute the loss.
loss.backward() # Derive gradients.
optimizer.step() # Update parameters based on gradients.
optimizer.zero_grad() # Clear gradients.
def test(loader):
model.eval()
correct = 0
for data in loader: # Iterate in batches over the training/test dataset.
out = model(data.x.float(), data.edge_index, data.batch)
pred = out.argmax(dim=1) # Use the class with highest probability.
num_batch = pred.shape[0]
correct += int((pred == data.y[:num_batch, 0]).sum()) # Check against ground-truth labels.
return correct / len(loader.dataset) # Derive ratio of correct predictions.
R = 10 # number of epochs
for epoch in range(0, R):
train()
train_acc = test(train_loader)
test_acc = test(test_loader)
print(f'Epoch: {epoch:03d}, Train Acc: {train_acc:.4f}, Test Acc: {test_acc:.4f}')
Epoch: 000, Train Acc: 0.9342, Test Acc: 0.9470
Epoch: 001, Train Acc: 0.9342, Test Acc: 0.9470
Epoch: 002, Train Acc: 0.9342, Test Acc: 0.9470
Epoch: 003, Train Acc: 0.9342, Test Acc: 0.9470
Epoch: 004, Train Acc: 0.9342, Test Acc: 0.9470
Epoch: 005, Train Acc: 0.9342, Test Acc: 0.9470
Epoch: 006, Train Acc: 0.9342, Test Acc: 0.9470
Epoch: 007, Train Acc: 0.9342, Test Acc: 0.9470
Epoch: 008, Train Acc: 0.9342, Test Acc: 0.9470
Epoch: 009, Train Acc: 0.9342, Test Acc: 0.9470
